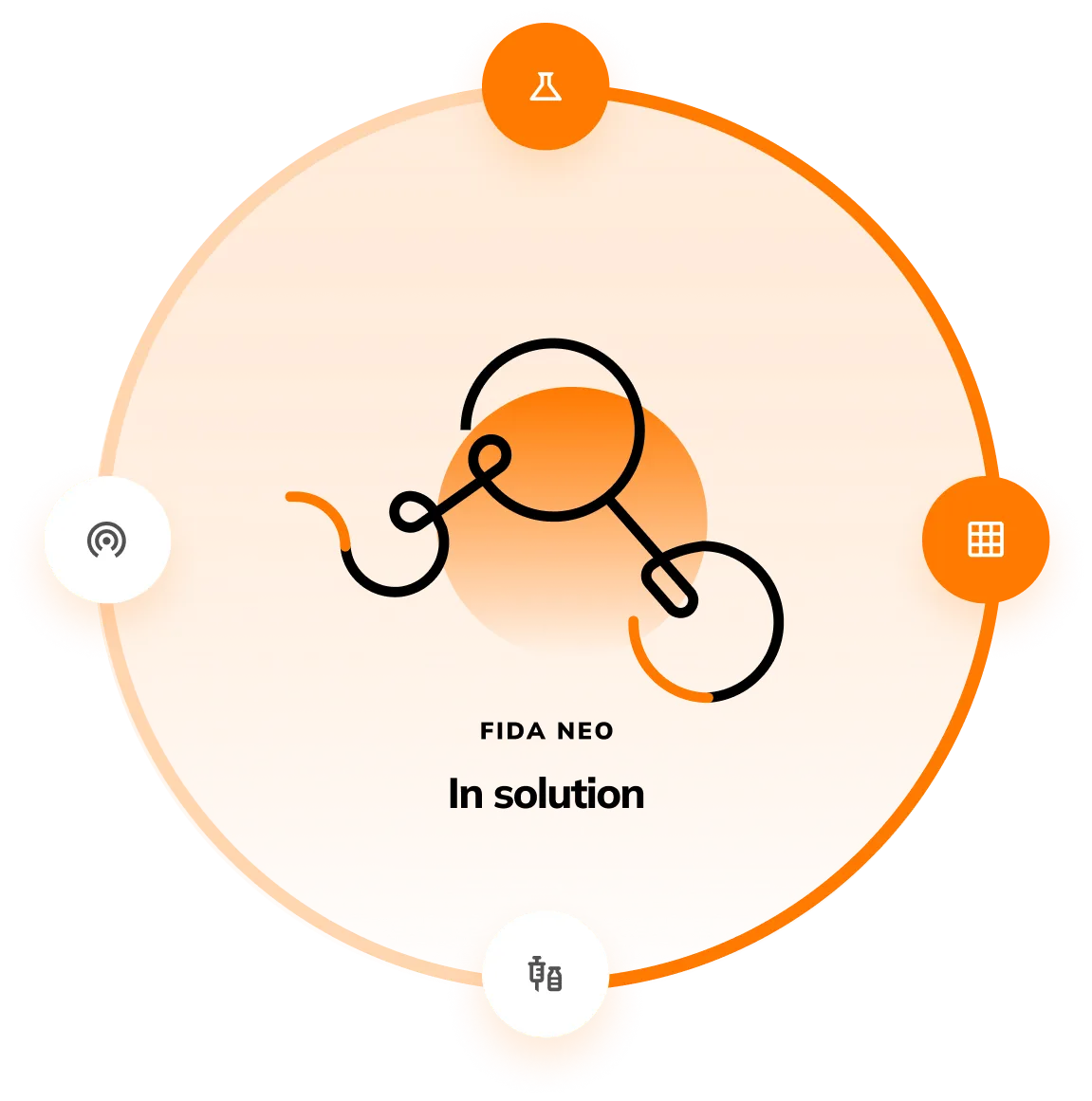
Fida Neo
Increasing Efficiency
with Biophysics
with Biophysics
Over 10 data points in one assay
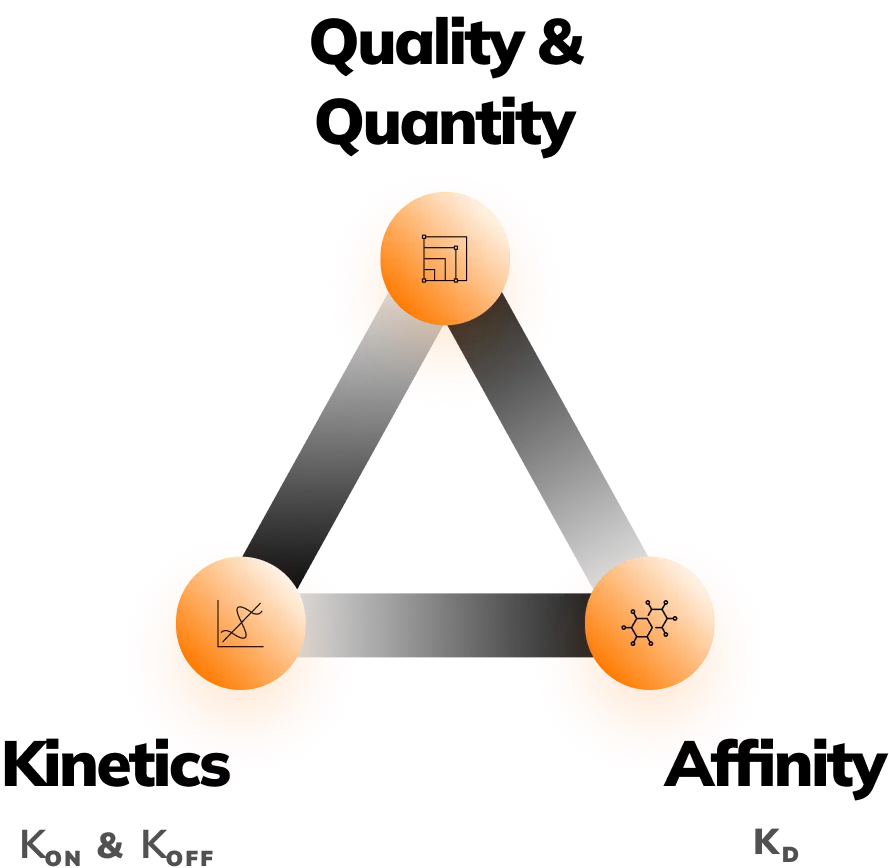
Measure: Affinity, kinetics, quantity & quality
Fida Neo delivers:
- Absolute size measurement (Rh)
- BRIC (Binding Related Intensity Change measurement)
Affinity (KD)
Kinetics (KON & KOFF)
- Quantity
Providing you with extensive data set and research flexibility.
All based on first principle, absolute measurements. Read more on technology
Benefit from 8 Quantitative Quality Control Parameters
8 embedded quality control features and a brand new Quality Control Module, deliver an overview on quality of each sample tested. Thanks to our newest update you can easily create A Quality Control Report to each of your essays, ensuring data consistency, boosting efficiency. Easily implemented in your workflow.

Used absolute size measurement (Rh)
Use FIDA to discover what triggers changes of your molecule or construct, based on direct measurement of the hydrodynamic radius. It detects size changes of less than 5% in hydrodynamic radius, has a dynamic size ranging from 0.5 to 500nm Rh and can detect affinities from pM to mM. These features make Fida Neo a perfect tool for affinity assays, as well as stoichiometry validation and oligometric/structural state of proteins
5%
size change detection
0.5-500nm
Dynamic Range
pM-mM
Affinities
sec-hrs
kinetics

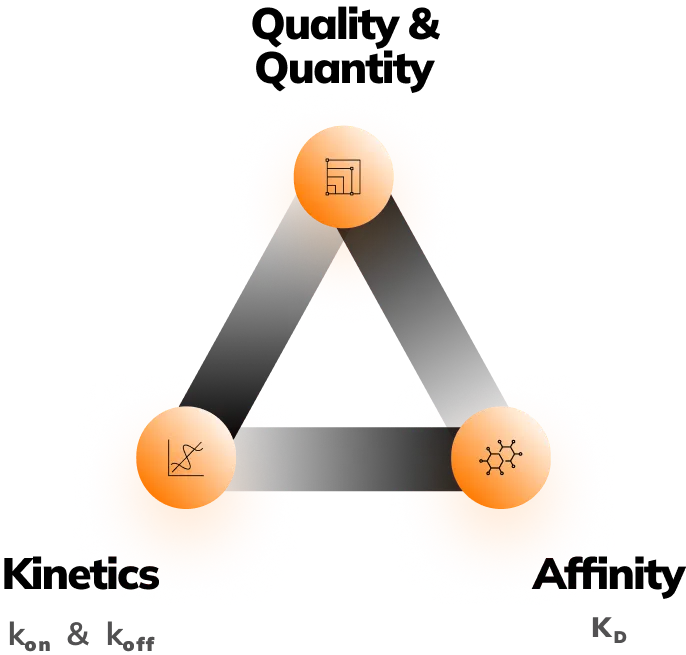
Measure: Affinity, kinetics,
quantity & quality
Fida Neo delivers 3 simultaneous measurements:
- Absolute size measurement (Rh)
- Lambda Dynamics (fluorescent ratiometric measurement)
- BRIC (Binding Related Intensity Change measurement)
Making it suitable for studying Affinity (KD), Kinetics (kon & koff ), Quantification & Sample Charaterization. Providing you with research flexibility and analytical freedom.
Read more on technology.
Read more on technology.
Benefit from 8 Quantitative Quality
Control Parameters
8 embedded quality control features and a brand new Quality Control Module, deliver an overview on quality of each sample tested. Thanks to our newest update you can easily create A Quality Control Report to each of your essays, ensuring data consistency, boosting efficiency. Easily implemented in your workflow.


Use absolute size measurement (Rh)
Use FIDA to discover what triggers changes of your molecule or construct, based on direct measurement of the hydrodynamic radius. It detects size changes of less than 5% in hydrodynamic radius, has a dynamic size ranging from 0.5 to 500nm Rh and can detect affinities from pM to mM. These features make Fida Neo a perfect tool for affinity assays, as well as stoichiometry validation and oligometric/structural state of proteins
1%
size change detection
0.5-500nm
Dynamic Range
pM-mM
Affinities
sec-hrs
kinetics
Work in-solution:
with no environmental restrictions
Fida Neo allows users to work under native conditions, such as serum, plasma, cell lysate or fermentation media. Having no restrictions on the matrix type unlocks the opportunity to run assays in environmentally relevant conditions.
- Seamlessly operate in complex matrices including fermentation media, plasma or serum.
Work in solution:
avoid non-specific binding
FIDA, as an non-surfaced, in solution technology keeps all the binding sites available for interaction. In practice, such an approach translates to:
- No steric hindrance to high density immobilised ligands
- No non-specific binding issues
- No risk of re-binding
Free yourself from restrictions:
any detergents, ionic strengths, temperature, pH and others
Fida Neo has no restrictions related to detergent usage, ionic strength, temperature or sample pH. This eliminates common research restrictions, boosts creative biophysical capability and allows you easily to produce environmentally relevant findings.
- Minimise assay development time
- Expand the scope of biological systems you can characterise
- Increase environmental relevance
No need for surface regeneration
With FIDA there is no surface chemistry involved.
- Eliminate risk of denaturing immobilised protein
- Rapidly determine slow off rates for high affinity interactions

An autosampler
2 x 96 well plates & 2 x 50 vials
2 x 96 well plates & 2 x 50 vials

1
Wide sensitivity range: picomolar to millimolar

3
Temperature controls:
- Capillary chamber
15-55°C - Autosampler
5-55°C

2
Labelled & Label free assays with:
480 nm, 640 nm or 280 nm LED detectors

4
1
An autosampler
2 x 96 well plates & 2 x 50 vials
3
Wide sensitivity range: picomolar to millimolar
2
Temperature controls:
- Autosampler 5-55
- Capillary chamber 10-45
4
Labelled & Label free assays with:
480 nm, 535 nm, 640nm or 280nm LED detectors








.webp)




Free yourself

No immobilisationIn-solution nature of FIDA allows for access to all binding sites - no more non-specific binding issues.

No constrainsCrude or purified samples, any pH, ionic strength limits, detergents or buffers.

No regenerationEliminates risk of denaturing immobilised protein. Allows for fast determination of slow off rates for high affinity interactions.
Stay in control

Flexible Assay DesignAdjust interaction times for kon/koff measurement; modulate mixing time through in-capillary sample mobilisation.

Embedded Quality Control ReportingFull transparency of sample material quality thanks to embedded Quality Control Module & Reporting Tool.

Detect Strong & Weak BindersCapable of measuring kinetics of both strong and weak interactions in-solution.
Boost Efficiency

Small sample volumes With as little as 4 uL analyte with fixed 40 nL indicator. Save material & effort.

No purificationMinimise assay development time and expands the range of possible biological systems to be characterised.

No time wastedRun 3 minute long assays & take informed decisions thanks to high data transparency.

Label-free or labelledHave an an option of switching detectors while using a single base instrument.

No expert requirementsWith just a few hour of training all scientists can run FIDA experiments.

Small sample volumes With as little as 4 uL analyte with fixed 40 nL indicator. Save material & effort.

No purificationMinimise assay development time and expands the range of possible biological systems to be characterised.

No time wastedRun 4 minute long assays & take informed decisions thanks to high data transparency.

Label-free or labelledHave an an option of switching detectors while using a single base instrument.

No expert requirementsWith just a few hour of training all scientists can run FIDA experiments.
Quality Control Reportingfor every sample


.png)


.webp)




How FIDA works, explained.



Learn More about FIDA Technology
Your laboratory instruments should serve you, not the other way around. We're happy to help you.
Who uses Fida?
Who uses Fida?