How is FIDA used in computational protein design?
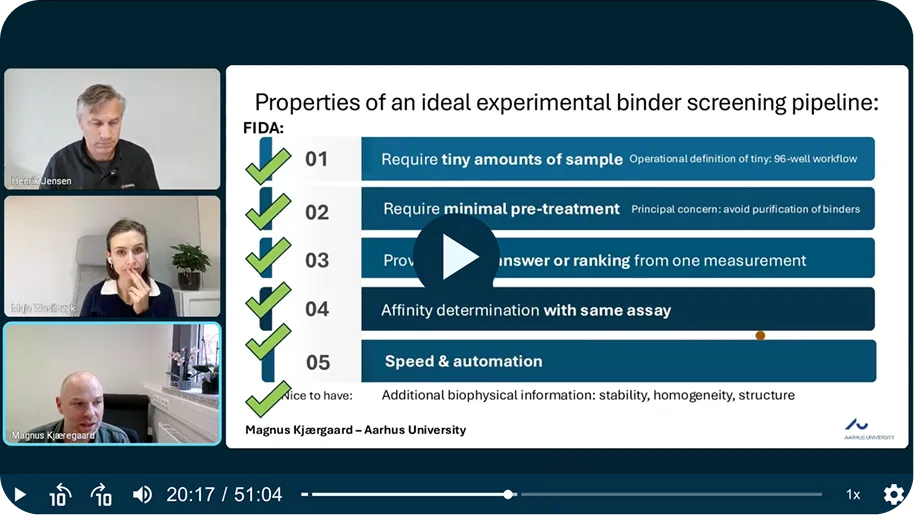
01
Direct in-lysate screening of synthetic proteins
- Analyze binders directly in unpurified or heat-treated lysate (or complex media).
- No purification, or immobilization required.
- Maintains native interaction conditions.
02
Quantitative affinity & size measurement
- One platform delivers: binding screening, Kd, and aggregation measurements.
- No false-positives, thanks to first-principle measurements.
- Robust: Absolute, quantitative data.
03
High sensitivity, low
sample use
- Highly sensitive: Detect small differences between similar de novo mini-binders.
- Down to 40 nL volumes.
- 96-well plate workflows.
04
Workflow-ready for binder development
- Screen, rank, and characterize your de novo binders in one workflow.
- Provides reproducible, biophysical readouts.
- Easy pipeline integration (straighforward assay development).
05
In-solution biophysics for challenging targets
- Suitable for dynamic, unstructured, or novel protein scaffolds.
- Measures interactions in solution (no surface artefacts).
- Includes stability, homogeneity, and structure information.
Bridging design and discovery
FIDA speeds up de novo binder development by removing purification and assay adaptation steps. With FIDA binders can be screened and ranked (Kd) directly in lysate, using only microliter sample volumes. The FIDA data is absolute and quantitative, allowing fast, side-by-side comparison of designed variants under native conditions, which proves especially useful in de novo mini-binder protein screening.

Learn more during an exploratory call with Fidabio team.
We are happy to answer all of your questions. Book an exploratory call to learn more about FIDA and the match between your personal needs and what we can deliver. The call is non-binding and free of any charges, so feel free to fill the form!