Supporting the Next Wave of Protein Design. Using FIDA for de novo binders
A practical challenge in de novo and computational protein design is that many targets (especially intrinsically disordered regions) lack tool compounds, making experimental validation dependent on developing and testing new binders from scratch. Early wet-lab screening therefore becomes a critical bottleneck.
Flow Induced Dispersion Analysis (FIDA) is an in-solution biophysical method that quantifies both molecular size (hydrodynamic radius) and interactions with high sensitivity, using minimal sample and without purification or immobilization. It is based on Taylor dispersion and the Stokes–Einstein equation, allowing FIDA to resolve size changes <5%, which provides a first-principle, absolute foundation for affinity and stability measurements.
What’s the most important capacity expansion brought by FIDA to the AI Protein Design field?
First, Direct screening of binders in lysates
This is the single most relevant capability for computational protein design labs.
Most de novo workflows produce dozens to hundreds of minibinders with only small expression volumes available. Purification is often the largest throughput bottleneck.
FIDA removes this constraint by allowing researchers to:
- Measure binders directly in unpurified or heat-treated lysate.
- Maintain native interaction conditions.
- Avoid assay development, immobilization steps, and surface adaptation completely.
- Apply 96-well workflows with microliter-scale sample volumes.
This directly supports labs in a built-up phase where expression capacity, time and personnel are limiting.
A simple heat-lysis workflow is sufficient: break cells, inactivate proteases, remove most host proteins, and proceed directly to FIDA. This setup is already used in medium-throughput screening pipelines.
Second, FIDA allows you to get structural information included when you do your functional assessments (2-in-1)
Because FIDA measures hydrodynamic radius, researchers obtain not only binding and affinity but also structural and solution-state information:
- Conformational changes
- Stability and homogeneity
- Aggregation or self-association
- Effects of mutations or design variants on size and shape
All measured in the same assay as binding. FIDA also supports two measurement modes depending on the binder’s kinetic behaviour:
- Capillary-mix mode: guarantees equilibrium, ideal for slow binders.
- Pre-incubation mode: reduces handling, sample consumption, and non-specific binding.
This flexibility is well suited for designed binders where kinetics and stoichiometry are often unknown.
Case examples from screening workflows
ALFA-tag binders (purified → lysate)
A panel of designed ALFA-tag binders showed clear separation with FIDA:
3–4 strong binders (Kd < 10 µM) identified in purified protein, and the same discrimination reproduced directly in lysate, with no purification required.

PSD-95 GK domain (low ΔRh, challenging target)
The target (2–3× larger than the binders) gave only 8–20% expected Rh changes, yet FIDA could still rank the 12 design variants directly in 30× diluted heat lysate, identifying 3 positives that were later validated by purified Kd (44–727 nM). These examples highlight FIDA’s ability to resolve small size changes and maintain accuracy in complex sample matrices.
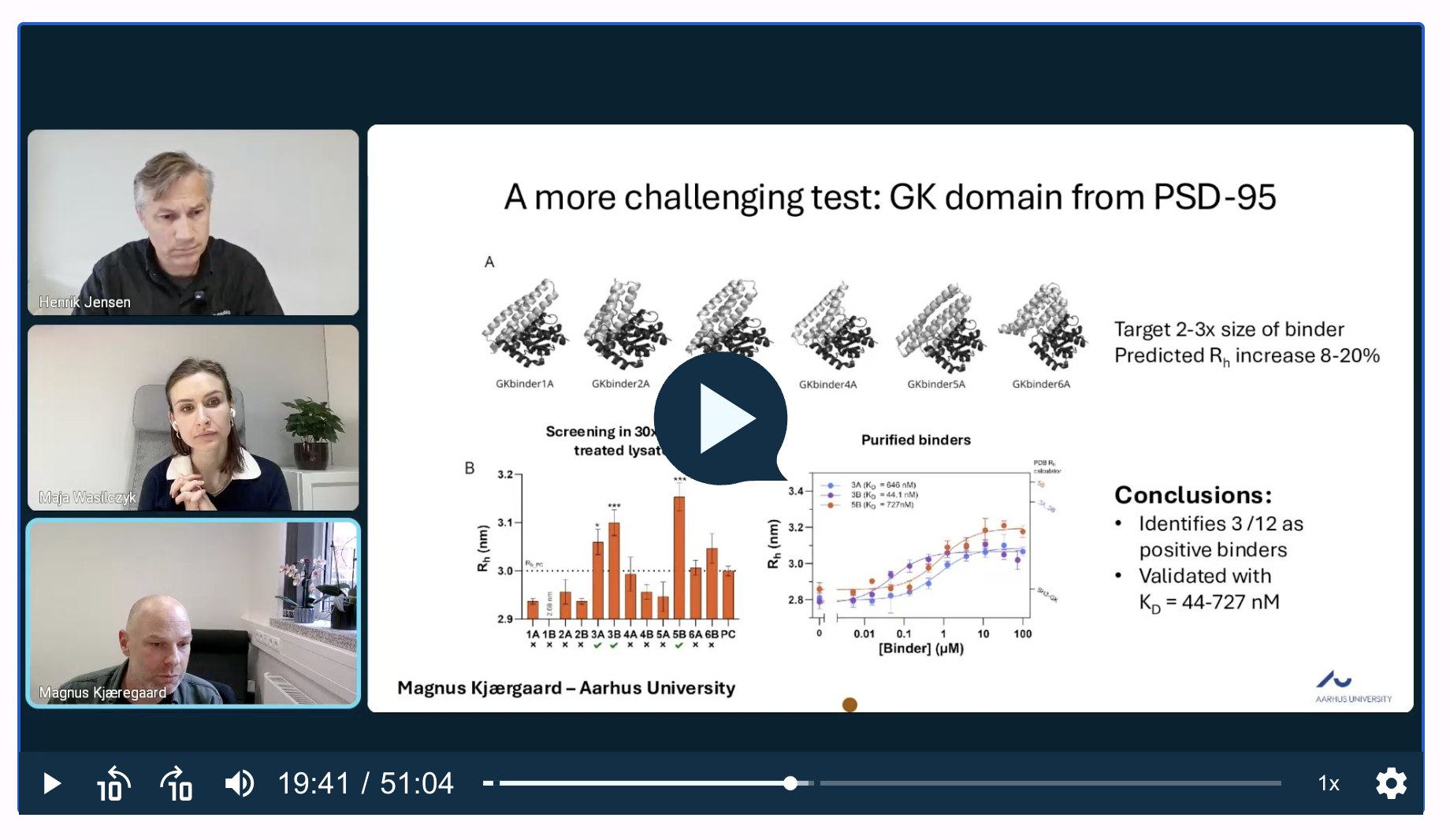
You can explore these cases by watching our on demand webinar here.
Metal-binding protein screening at scale
Beyond binder–target interactions, FIDA has also been applied to screen large panels of metal-binding proteins, where conformational changes upon metal coordination can be subtle or highly pronounced. In a recent workflow assessing 34 proteins across multiple metals, FIDA reliably detected interactions using any of its three orthogonal readouts: hydrodynamic radius, fluorescence intensity, or fluorescence ratio. Out of 34 proteins, 26 showed binding, and the same instrument was then used to run full titrations on selected hits using the same batch of protein. In a second campaign, a 95-mutant library was screened for improved Mg²⁺ affinity, with 50 mutants outperforming wild-type and top candidates achieving more than 30-fold affinity improvement, all identified, filtered for quality, and characterized within one week.

Bridging design and discovery
FIDA accelerates de novo binder development by removing purification and assay-adaptation steps altogether. Binders can be screened and ranked directly in lysate using only microliter volumes. The resulting data are absolute and quantitative, enabling fast, side-by-side comparison of designed variants under native conditions.
This makes FIDA particularly well suited for rapidly scaling AI/protein design labs, where the bottleneck is no longer computational creativity but experimental validation capacity. Learn more during a 1:1 call.
.png)

.png)